
Optogenetics involves the combined use of light with genetically engineered modules to control cellular and molecular processes. The Tucker Lab works to engineer new protein-based tools to control protein interactions and protein activity with light. To accomplish this, we reengineer photosensory domains from light-sensitive organisms such as plants to control activity of other proteins in new ways. As light is an ideal actuator, light-regulated systems allow spatial, temporal, dose-dependent, and reversible control.
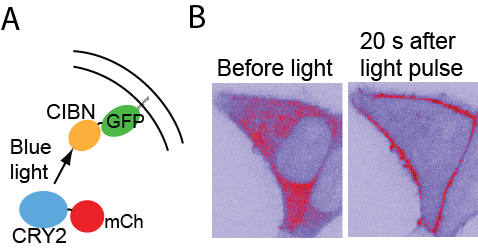
Control of protein-protein interactions with light using CRY2/ CIB1 system. (A) Cytosolic CRY2-mCherry is recruited to a membrane-localized CIB1 fragment upon light application. (B) Localization of CRY2-mCherry before and 20s after recruitment.
Cryptochrome 2-based optogenetic tools
We pioneered use of the plant cryptochrome protein CRY2, along with a light-dependent interactor, CIB1, to generate the CRY2/CIB1 photodimerizer system. This system allows fast and local regulation of protein localization and/or activity using light.
See: Kennedy et al., 2010 Nature Methods
PA-BoNT
Botulinum toxin is a protease that targets SNARE proteins involved in synaptic release of neurotransmitters. In collaboration with the Kennedy Lab, we developed a split botulinum toxin B that can be reconstituted with light. Thus, light triggers protease activity, resulting in cleavage of the SNARE target (VAMP-2) and blockage of neurotransmitter release.
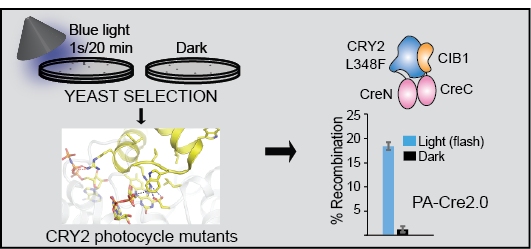
PA-Cre2.0
We used yeast directed evolution to engineer a second-generation photoactivatable Cre DNA recombinase, in which activity is induced with light. A split recombinase enzyme is is brought together by CRY2-CIB1 interaction, reconstituting activity in light.
See Taslimi et al, 2016 Nature Chemical Biology
Also: Meador et al., 2019 Nucleic Acids Research
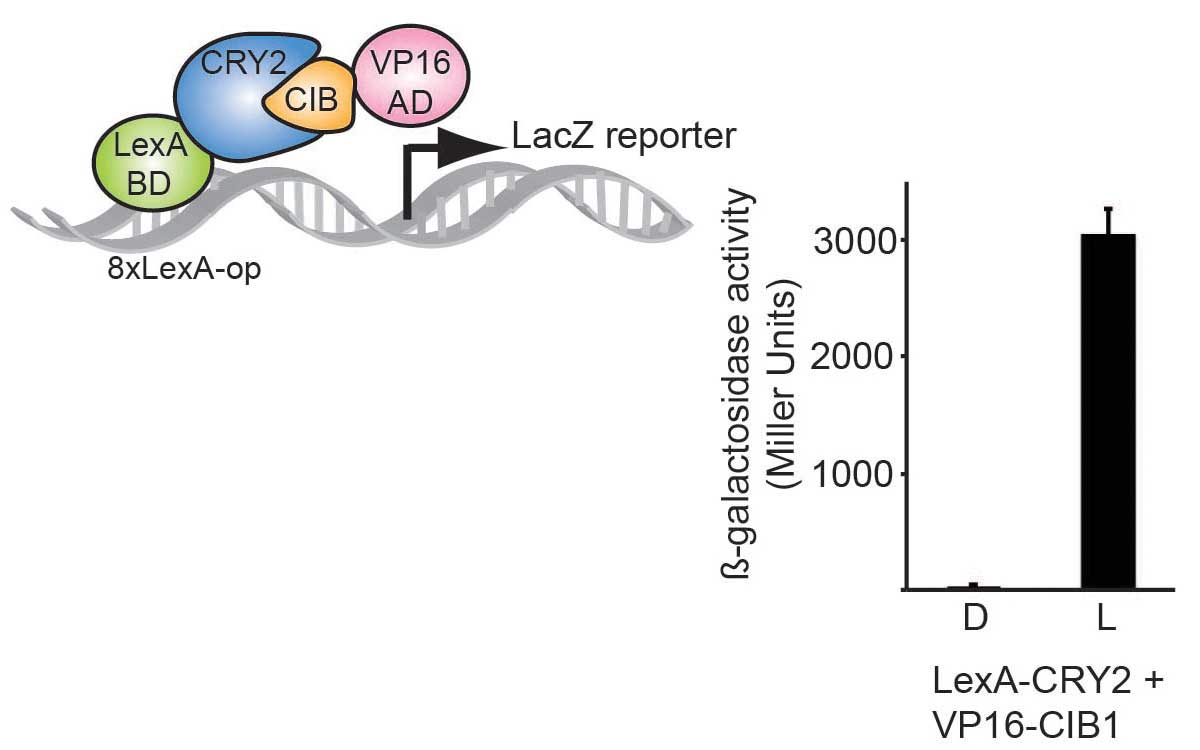
We used the CRY2-CIB1 interaction to reconstitute a split Gal4BD-VP16AD transcription factor, allowing robust induction of transcription with light.
We also developed a method to block transcription factor activity with light in mammalian cells.
See Pathak et al., 2014; Hughes et al., 2012; Kennedy et al., 2010; Pathak et al., 2017
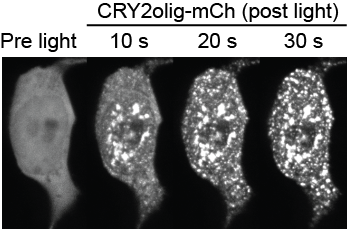
CRY2olig
We developed a new optogenetic tool, CRY2olig, that allows robust and reversible protein clustering with light. In the figure at left, a blue light pulse (488 nm) applied to cells expressing CRY2olig-mCherry causes redistribution of protein into large clusters.
See: Taslimi et al., 2014 Nature Communications
Manipulation of protein activity using CRY2olig condensates/clusters
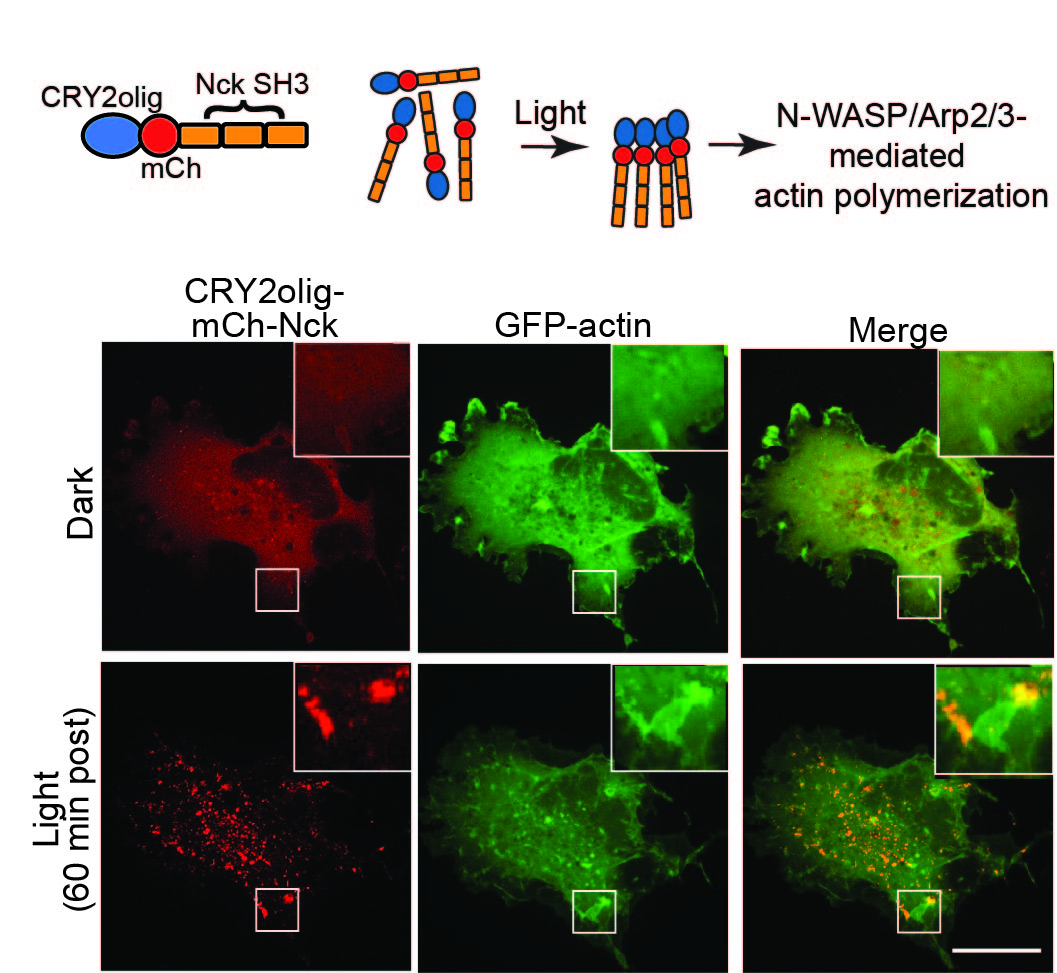
We are exploring use of CRY2olig to disrupt and/or activate target protein function and to visualize protein-protein interactions. At left, we used CRY2olig to trigger clustering of Nck SH3 domains with light, resulting in actin clustering and perturbations to the cytoskeleton.
See: Taslimi et al., 2014 Nature Communications
In addition, by fusing CRY2olig to different protein targets, we can form protein condensates. We are interested in understanding the regulation of and cellular response to protein condensate formation.
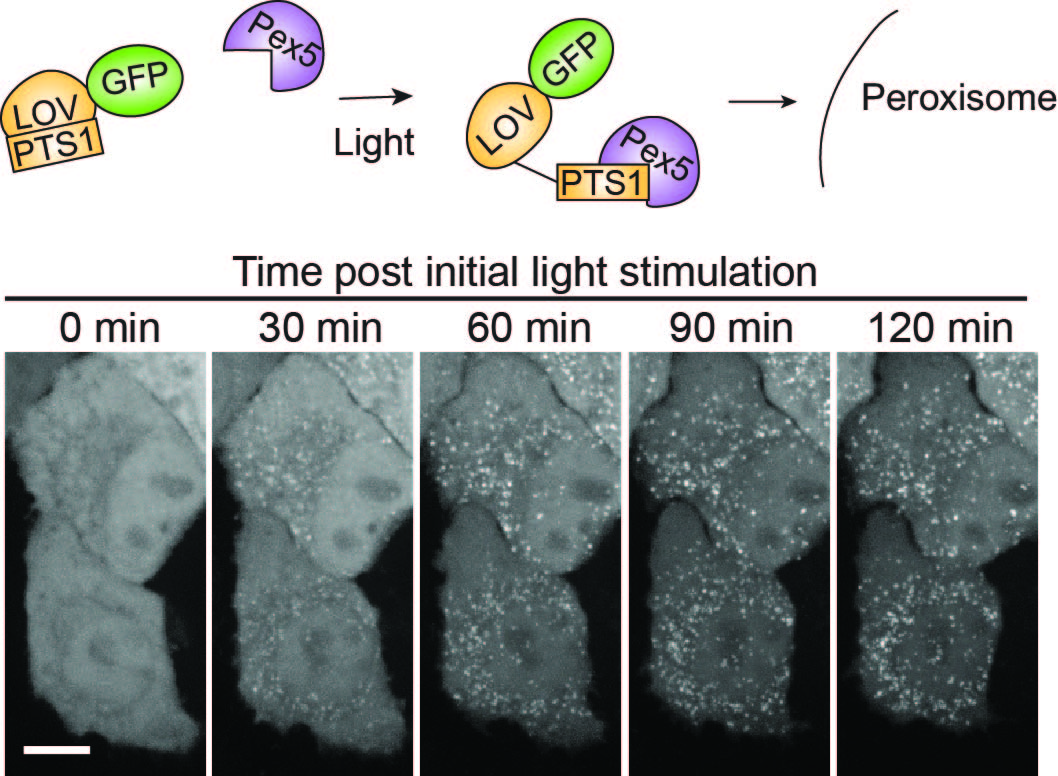
We are using optogenetic approaches to control protein trafficking and other cellular events. In this figure, we controlled trafficking of peroxisomal targets with light using AsLOV2. Cells expressing GFP-LOV-PTS1 are primarily cytosolic in dark, but light triggers exposure of a PTS1 tag that binds Pex5, the peroxisome import receptor, resulting in peroxisomal import and depletion from the cytosol.
See: Spiltoir et al., 2015 ACS Synthetic Biology
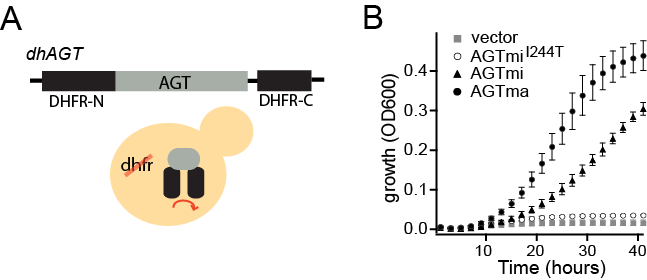
DHFR protein misfolding reporter
We are interested in yeast cell-based reporters for protein misfolding. One such reporter we developed uses a target protein inserted within the essential enzyme dihydrofolate reductase (DHFR). Using this reporter in a DHFR-deleted yeast strain (A), we obtain differences in growth of yeast that reflect differences in protein stability of disease variants (B).
See: Pittman et al., 2012
The protein alanine : glyoxylate aminotransferase (AGT) is a critical enzyme in both humans and yeast. In humans, deficiency leads to the disease primary hyperoxaluria type I, a severe kidney disease. Many disease-linked mutations have been shown to destabilize AGT, leading to misfolding and subsequent degradation or mislocalization. We have generated 'humanized' yeast that contain human AGT in place of the yeast version of alanine:glyoxylate aminotransferase, and have used this approach to further characterize the human disease protein.
See: Lage et al., 2014; Pittman et al., 2012; Hopper et al., 2008
